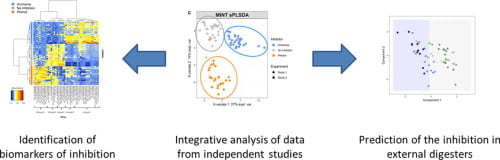
Integrating independent microbial studies to build predictive models of anaerobic digestion inhibition by ammonia and phenol
Our collaboration with A/Prof Olivier Chapleur lab (INRAE, France) continues in anaerobic digestion!
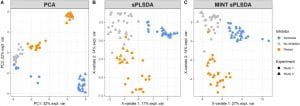
In this study we applied our MINT framework* to integrate independent 16S studies to identify microbial indicators of digesters inhibition, and ultimately aim to improve anaerobic digestion management. We trained our MINT model on two in-house studies, then predicted ammonia inhibition in two external and published studies with 90% accuracy.
Key highlights of the study:
- Robust biomarkers of AD inhibition by ammonia and phenol were tagged by integrating independent 16S studies.
- Multivariate model predicts ammonia inhibition with 90% accuracy in 2 external studies.
- Increase of the Clostridiales relative abundance is an early warning of AD inhibition.
- Presence of Cloacimonetes is associated with good performance of methane production.
Reference:
Poirier S, Déjean, S, Midoux, C, Lê Cao K-A, Chapleur O (2020). Integrating independent microbial studies to build predictive models of anaerobic digestion inhibition by ammonia and phenol. Bioresource Technology” https://www.sciencedirect.com/science/article/pii/S0960852420312244
*MINT has been also been successfully applied to single cell RNA-seq studies:
- Al’s GitHub page: https://github.com/ajabadi/MINT_sPLSDA
- Tian L, Dong X, Freytag S, Lê Cao K-A, Su S, Abadi AJ, Amann-Zalcenstein D, Weber TS, Seidi S, Jabbari JS, Naik S, Ritchie ME (2019). Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nature methods 16(6):479-487.
- Rohart F, Eslami A, Matigian, N, Bougeard S, Lê Cao K-A (2017). MINT: A multivariate integrative approach to identify a reproducible biomarker signature across multiple experiments and platforms. BMC Bioinformatics 18:128.
Categories