New framework: timeOmics published
In collaboration with Antoine Bodein and Arnaud Droit from Université de Laval (bioinformatics) and Olivier Chapleur (IRSTEA, France, microbiology), we have developed the timeOmics framework to handle time course and longitudinal datasets. The paper is now published, along with our gitHub page repo in progress.
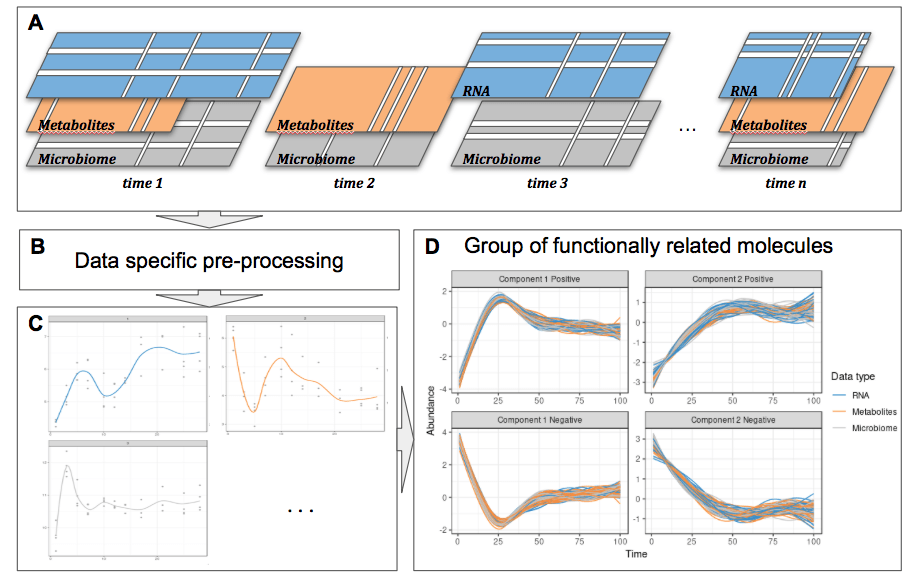
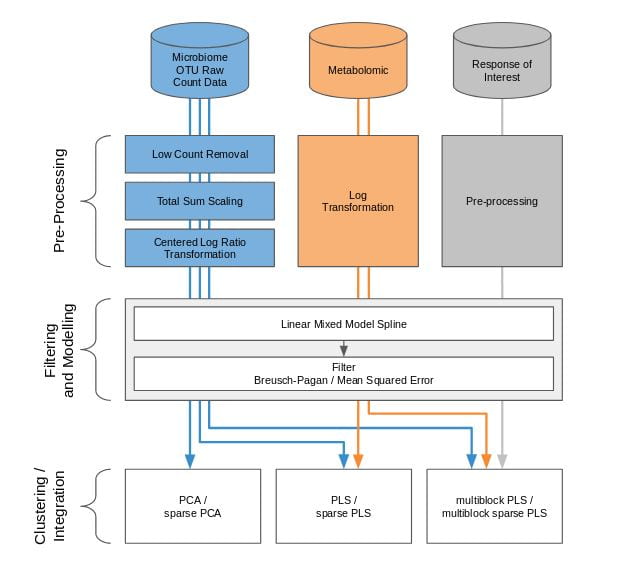
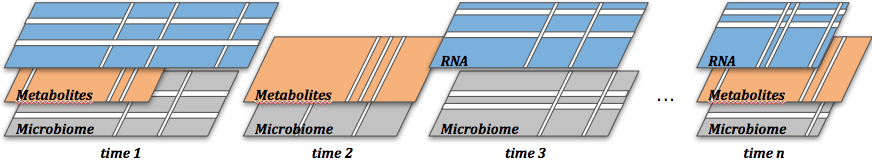
We have applied our framework in two microbiome related studies.
The gut infant microbiome development study investigates the stool microbiome of 11 infants during their first year of life. The purpose of our statistical analysis was to identify a bacterial signature that describes the dynamics of a baby’s microbial gut development in the first days of life, as well as compare differences in signatures between babies born by vaginal delivery or by C-section. As this study is single omics, we applied our framework depicted in the figure above with sparse PCA.
The waste degradation study investigates anaerobic digestion (AD) in three lab-scale bioreactors across 150 days (4 or 5 time points). AD involves a complex microbiome that is responsible for the progressive degradation of molecules into methane and carbon dioxide. The purpose of the study was to investigate the relationship between biowaste degradation performance, microbial and metabolomic dynamics across time. The aim of our statistical analysis was to identify highly associated multi-omic signatures characterizing waste degradation dynamics in the 3 bioreactors. This study involves the integration of two omics datasets and degradation performance measures, we applied sparse PLS and multiblock s[arse PLS, as shown in the figure above.
Categories