A simple, scalable approach to building a cross-platform transcriptome atlas
A fruitful result from our long standing collaboration with Prof Christine Wells’ group at the Centre for Stem cell Systems . Our approach enables to build transcriptome atlas across several RNA-seq and microarray platforms. There is also a possibility to project single cell RNA-seq on the atlas itself to bring more insights into the biology of cells!
This is the first manuscript of our Honours student Yidi Deng, co-supervised by Jarny Choi, Christine Wells and myself. Well done! 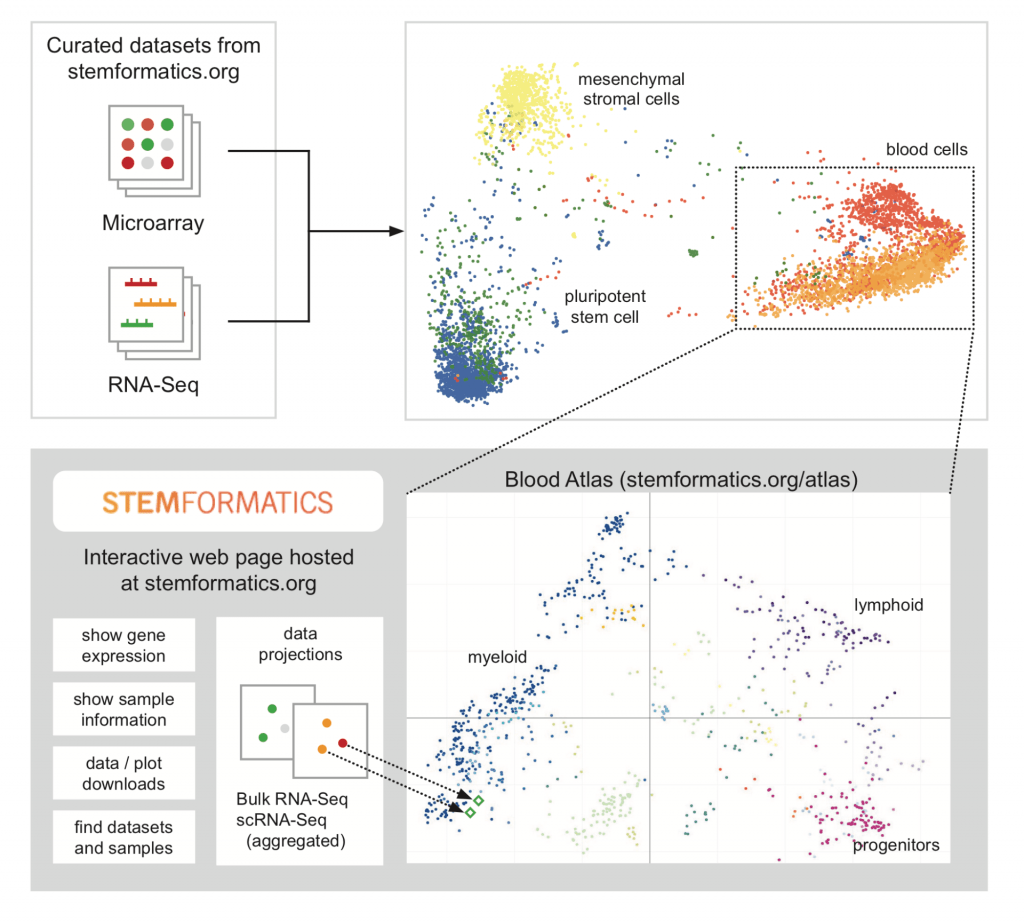
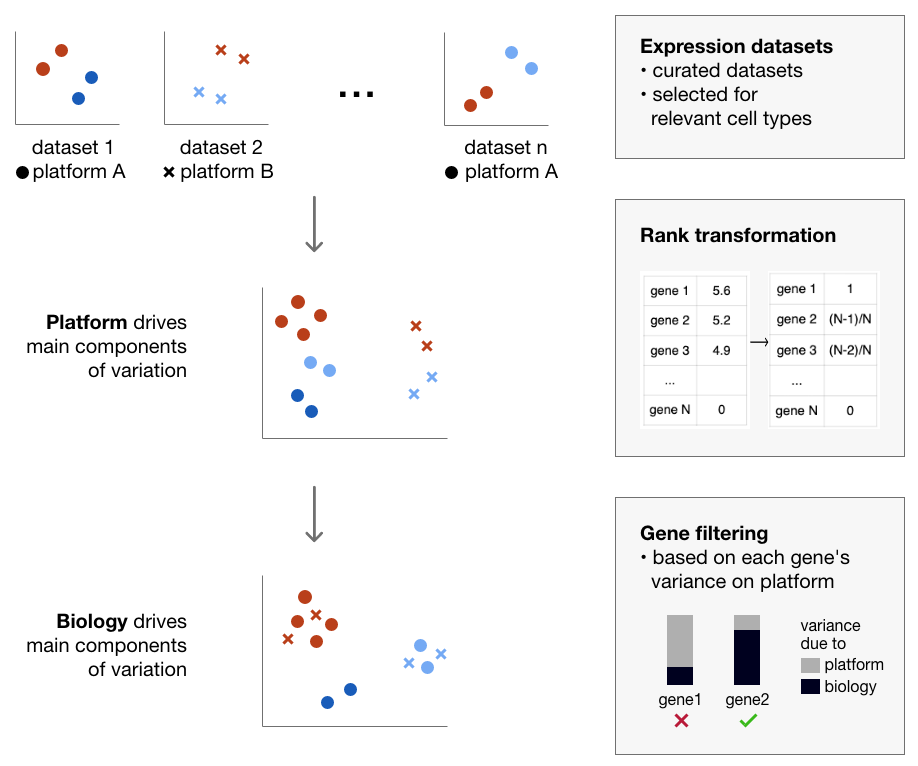
Combining data from many different studies is an attractive way of capturing new aspects of the biology being studied. Biological variance attributable to cell type, cellular niche, origin, disease status or environmental stimuli is the basis of most small-n transcriptome studies. In aggregation, these promise to capture emergent dimensions of a biology that is not possible to view from any individual study. However biological signal is easily swamped by technical artifact, especially when data is generated on platforms with profoundly different data structures. This is the case when comparing microarray data to RNAseq, or RNAseq to single cell profiling. Consequently, transcriptome atlases are generally comprised from a small number of donors/conditions surveyed using one technology platform.
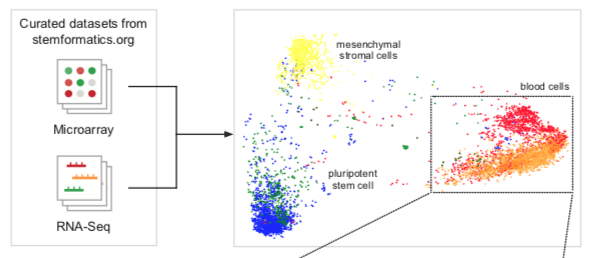
In this paper we present a simple and scalable data integration method that is platform agnostic. We provide a proof-of-principle by constructing an atlas of blood cells that combines many data sets measured on different platforms, and that in combination, recapitulates the known blood hierarchy. The atlas provides a reference to compare external samples to, allowing users to benchmark new derivation or isolation methods. It also provides a reference point for new data types, such as the classification of single cells. The approach allows for FAIR data reuse and robust identification of molecular signatures across multiple studies and experimental conditions.
Angel PW, Rajab N, Deng Y, Pacheco CM, Chen T, Lê Cao K-A, Choi J, Wells CA (2020). A simple, scalable approach to building a cross-platform transcriptome atlas. PLoS Computational Biology (current version is on bioRxiv)
Categories