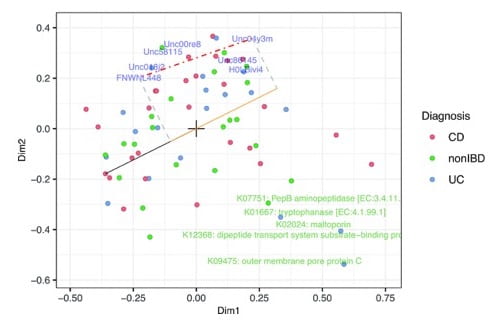
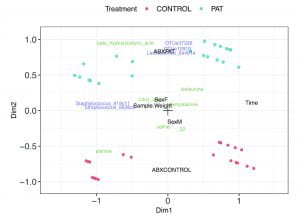
Model-based joint visualization of multiple compositional omics datasets
Stijn Hawinkel did a 4 month PhD placement in our lab in 2019 and developed a neat approach to integrate compositional multi-omics data sets.
Stijn introduces.a new latent variable model to integrate and visualise multi-omics data, while handling covariates, missing values, compositional structure and heteroscedasticity. The compositional biplots enable to represent the different data views. The approach was benchmarked on three multi-omics studies, ranging from host transcriptomics, metabolomics, proteomics, microbiome, meta-proteome.
The algorithm is available in the R bioconductor package combi.
Reference:
Hawinkel S, Bijnens L, Lê Cao K-A, Olivier Thas (2020). Model-based joint visualization of multiple compositional omics datasets, NAR Genomics and Bioinformatics
Categories