Posts tagged with Manuscript
-
CellDiffusion: a generative model to annotate single-cell and spatial RNA-seq using bulk references

We introduce the first manuscript of our PhD student Xiaochen Zhang! CellDiffusion bridges the gap between single-cell, spatial and bulk RNA-seq, using a diffusion-based generative model to create virtual cells that mimic the gene expression profiles …
lecao-lab.science.unimelb.edu.au/2025/10/30/celldiffusion-a-generative-model-to-annotate-single-cell-and-spatial-rna-seq-using-bulk-references
-
PLSKO: a robust knockoff generator to control false discovery rate in omics variable selection

Our PhD student Guannan Yang has submitted her first manuscript! PLSKO is a new and robust knockoff variable generator that is applicable to various types of …
lecao-lab.science.unimelb.edu.au/2024/08/15/plsko
-
StableMate to select stable predictors in omics data
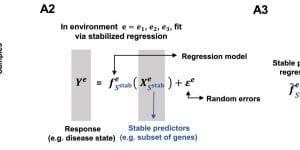
Variable selection in ‘omics data is essential to identify reliable markers of diseases, or shed light into molecular mechanisms. however, many variable selection methods in multivariate analysis (e.g. …
lecao-lab.science.unimelb.edu.au/2023/10/19/stablemate
-
Metaphor: A workflow for streamlined assembly and binning of metagenomes
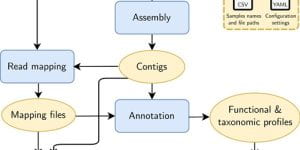
Genome-resolved metagenomics techniques aim to recover genomes from high-throughput sequencing data, and have led to unprecedented insight into microbial diversity, ecology, and evolution. However, data pipelines …
lecao-lab.science.unimelb.edu.au/2023/08/02/metaphor
-
A simple, scalable approach to building a cross-platform transcriptome atlas
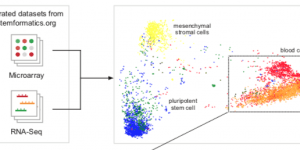
A fruitful result from our long standing collaboration with Prof Christine Wells’ group at the Centre for Stem cell Systems . Our approach enables to build transcriptome …
lecao-lab.science.unimelb.edu.au/2020/09/08/a-simple-scalable-approach-to-building-a-cross-platform-transcriptome-atlas
-
Article published: Variable selection in microbiome compositional data analysis
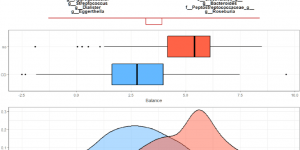
Antoni Susin, Yiwen Wang, Kim-Anh Lê Cao, M Luz Calle (2020) Variable selection in microbiome compositional data analysisNAR Genomics and Bioinformatics, Volume 2, Issue 2, June 2020, lqaa029, …
lecao-lab.science.unimelb.edu.au/2020/06/03/variable-selection-in-microbiome-compositional-data-analysis
-
Single cell benchmark manuscript on bioRxiv

The CellBench data are out and we benchmarked a wide variety of methods for single cell transcriptomics data analysis. More details at this link. This work …
lecao-lab.science.unimelb.edu.au/2018/10/04/single-cell-benchmark-manuscript-on-biorxiv

