Posted under single cell
-
CellDiffusion: a generative model to annotate single-cell and spatial RNA-seq using bulk references

We introduce the first manuscript of our PhD student Xiaochen Zhang! CellDiffusion bridges the gap between single-cell, spatial and bulk RNA-seq, using a diffusion-based generative model to create virtual cells that mimic the gene expression profiles …
lecao-lab.science.unimelb.edu.au/2025/10/30/celldiffusion-a-generative-model-to-annotate-single-cell-and-spatial-rna-seq-using-bulk-references
-
Φ-Space: Continuous phenotyping of single-cell multi-omics data with video!

Jiadong Mao has developed a new PLS method for cell type continuous annotation of single cells, now published in Genome Biology! Φ-Space addresses numerous challenges faced …
lecao-lab.science.unimelb.edu.au/2024/07/03/%cf%86-space
-
Sincast: a computational framework to predict cell identities in single-cell transcriptomes using bulk atlases as references
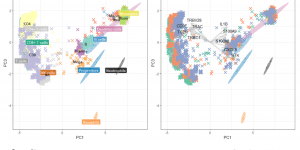
Following up from our preliminary work on how to query cell types from transcriptomics bulk studies using bulk atlases, our talented student Yidi Deng has gone …
lecao-lab.science.unimelb.edu.au/2022/04/03/sincast
-
Community-wide hackathons to identify central themes in single-cell multi-omics
Last year we ran an online event as part of Banff International Research Station workshops. We designed and analysed three single cell multi-omics hackathons with another …
lecao-lab.science.unimelb.edu.au/2021/08/06/community-wide-hackathons-to-identify-central-themes-in-single-cell-multi-omics
-
A simple, scalable approach to building a cross-platform transcriptome atlas
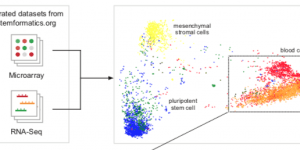
A fruitful result from our long standing collaboration with Prof Christine Wells’ group at the Centre for Stem cell Systems . Our approach enables to build transcriptome …
lecao-lab.science.unimelb.edu.au/2020/09/08/a-simple-scalable-approach-to-building-a-cross-platform-transcriptome-atlas