
Sincast: a computational framework to predict cell identities in single-cell transcriptomes using bulk atlases as references
Following up from our preliminary work on how to query cell types from transcriptomics bulk studies using bulk atlases, our talented student Yidi Deng has gone ahead to establish a new framework to query single cell transcriptomics studies based on transcriptomics bulk atlases!
Our article is now published:
Yidi Deng, Jarny Choi*, Kim-Anh Lê Cao*, Sincast: a computational framework to predict cell identities in single-cell transcriptomes using bulk atlases as references, Briefings in Bioinformatics, 2022;, bbac088, https://doi.org/10.1093/bib/bbac088
-
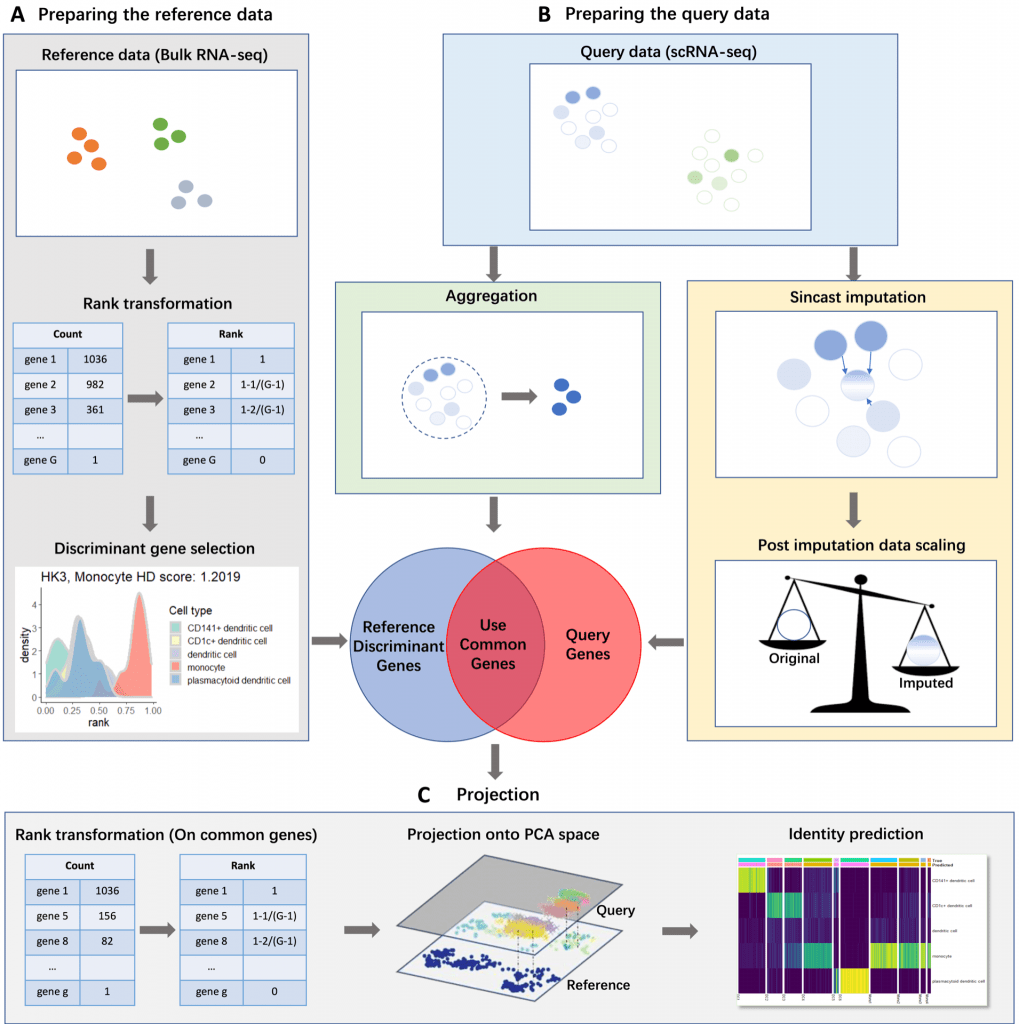
Sincast uses Rank Transformation and discriminant gene filtering based on Hellinger Distance to build the reference bulk RNA-seq atlases.
-
The query cells from scRNA-seq data can be either aggregated or zero imputed, without the need for batch effect correction.
-
Single cells are projected on the reference bulk atlas using PCA and diffusion mal allows visualization across several PC dimensions.
-
Cell prediction along a continuum allows to highlight new cell states.
-
Key gene regulators can be identified as well as pseudo-time trajectories.
Categories