Posted under microbiome
-
Semisynthetic Simulation for Microbiome Data Analysis

Dr Kris Sankaran (University of Wisconsin) was one of our Belz recipient from the School of Maths & Stats and came for 6-week visit to our …
lecao-lab.science.unimelb.edu.au/2025/01/20/semisynthetic-simulation-microbiome
-
Microbial network inference for longitudinal microbiome studies with LUPINE with video!

Dr Saritha Kodikara has developed a PLS-based to infer microbial networks across time! LUPINE is a PLS-based method that combines dimension reduction, and partial correlations to …
lecao-lab.science.unimelb.edu.au/2024/05/17/lupine
-
Metaphor: A workflow for streamlined assembly and binning of metagenomes
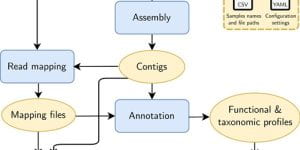
Genome-resolved metagenomics techniques aim to recover genomes from high-throughput sequencing data, and have led to unprecedented insight into microbial diversity, ecology, and evolution. However, data pipelines …
lecao-lab.science.unimelb.edu.au/2023/08/02/metaphor
-
Article: PLSDA-batch: a multivariate framework to correct for batch effects in microbiome data
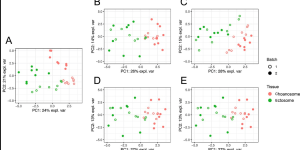
It’s been a long journey! The method developed by my former PhD student Eva Yiwen Wang is out and ready to be used! Yiwen Wang, Kim-Anh …
lecao-lab.science.unimelb.edu.au/2023/01/20/article-plsda-batch

