Posted under News
-
Translating big data into better health thanks to major grant

[from the faculty of science newsroom, written by Rebecca Colless] Biological markers predicting pre-eclampsia – a pregnancy disorder that causes premature births and serious health problems …
lecao-lab.science.unimelb.edu.au/2023/12/19/translating-big-data-into-better-health-thanks-to-major-grant
-
Dean’s Honours Award for PhD thesis to Dr Yiwen Wang

Our former student, Dr Yiwen (Eva) Wang was awarded the Dean’s Honours Award on November 22nd, an award that recognises excellence in a PhD thesis submitted …
lecao-lab.science.unimelb.edu.au/2023/12/04/deans-honours-award-for-phd-thesis-to-dr-yiwen-wang
-
StableMate to select stable predictors in omics data
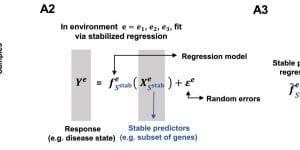
Variable selection in ‘omics data is essential to identify reliable markers of diseases, or shed light into molecular mechanisms. however, many variable selection methods in multivariate analysis (e.g. …
lecao-lab.science.unimelb.edu.au/2023/10/19/stablemate
-
mixOmics shortlisted in prestigious Eureka Prize from the Australian Museum

[from the University of Melbourne News room] Software developed by the Faculty of Science’s Professor Kim-Anh Lê Cao (School of Mathematics and Statistics) has been shortlisted …
lecao-lab.science.unimelb.edu.au/2023/07/27/mixomics-shortlisted-in-prestigious-eureka-prize-from-the-australian-museum
-
Self-paced online course May 22 – July 7 2023

Registrations for the third iteration of our online mixOmics course are now open! More details here. Feedback from our last iteration can be found here. If …
lecao-lab.science.unimelb.edu.au/2023/04/27/self-paced-online-mixomics-course-self-paced-online-course-may-2023
-
Article: PLSDA-batch: a multivariate framework to correct for batch effects in microbiome data
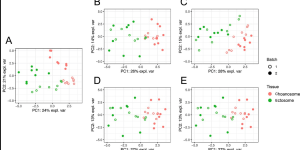
It’s been a long journey! The method developed by my former PhD student Eva Yiwen Wang is out and ready to be used! Yiwen Wang, Kim-Anh …
lecao-lab.science.unimelb.edu.au/2023/01/20/article-plsda-batch
-
Self-paced online mixOmics course Oct 31st – Nov 27 2022

Registrations for the second iteration of our online mixOmics course are now open! More details here.
lecao-lab.science.unimelb.edu.au/2022/08/14/self-paced-online-mixomics-course-oct-31st-nov-27-2022
-
Review article: Statistical challenges in longitudinal microbiome data analysis
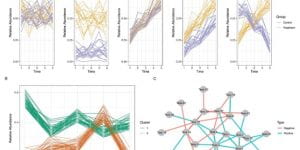
With Saritha Kodikara and Susan Ellul, we have published our latest review and benchmarked existing methods for longitudinal microbiome analysis on simulated data and a case …
lecao-lab.science.unimelb.edu.au/2022/07/27/longitudinal-microbiome-data-analysis
-
Sincast: a computational framework to predict cell identities in single-cell transcriptomes using bulk atlases as references
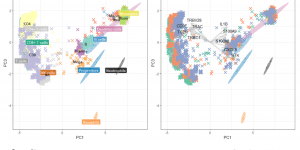
Following up from our preliminary work on how to query cell types from transcriptomics bulk studies using bulk atlases, our talented student Yidi Deng has gone …
lecao-lab.science.unimelb.edu.au/2022/04/03/sincast
-
Outreach: visit at Melbourne Girls’ College

With some the outreach team from the School of Mathematics and Statistics, I visited the Melbourne Girls’ College, a highly selective girls state school, but very …
lecao-lab.science.unimelb.edu.au/2022/02/28/outreach
Number of posts found: 62